max_depth is an interesting parameter. While n_estimators has a tradeoff between speed & score, max_depth has the possibility of improving both. By limiting the depth of your trees, you can reduce overfitting.
Unfortunately, deciding on upper & lower bounds is less than straightforward. It'll depend on your dataset. Luckily, I found a post on StackOverflow that had a link to a blog post that had a promising methodology.
First, we build a tree with default arguments and fit it to our data.
import pandas as pd
from sklearn.metrics import precision_recall_curve
from sklearn.metrics import auc
from sklearn.ensemble import RandomForestClassifier
from sklearn.datasets import load_breast_cancer
data = load_breast_cancer()
X, y = data.data, data.target
rfArgs = {
"random_state": 0,
"n_jobs": -1,
"class_weight": "balanced",
"n_estimators": 18,
"oob_score": True
}
clf = RandomForestClassifier(**rfArgs)
clf.fit(X, y)Now, let's see how deep the trees get when we don't impose any sort of max_depth. We'll use the code from that wonderful blog post to crawl our Random Forest, and get the height of every tree.
#From here: https://aysent.github.io/2015/11/08/random-forest-leaf-visualization.html
def leaf_depths(tree, node_id = 0):
'''
tree.children_left and tree.children_right store ids
of left and right chidren of a given node
'''
left_child = tree.children_left[node_id]
right_child = tree.children_right[node_id]
# If a given node is terminal, both left and right children are set to _tree.TREE_LEAF
if left_child == _tree.TREE_LEAF:
depths = np.array([0]) # Set depth of terminal nodes to 0
else:
# Get depths of left and right children and increment them by 1
left_depths = leaf_depths(tree, left_child) + 1
right_depths = leaf_depths(tree, right_child) + 1
depths = np.append(left_depths, right_depths)
return depths
allDepths = [leaf_depths(estimator.tree_)
for estimator in clf.estimators_]
print(np.hstack(allDepths).min())
print(np.hstack(allDepths).max())Here's the output:
#> 2
#> 9We'll be searching between 2 and 9!
Let's bring back our old make a helper function to easily return scores.
...
def getForestAccuracy(X, y, kwargs):
clf = RandomForestClassifier(**kwargs)
clf.fit(X, y)
y_pred = clf.oob_decision_function_[:, 1]
precision, recall, _ = precision_recall_curve(y, y_pred)
return auc(recall, precision)Now let's see it in action:
max_depth = bgs.compareValsBaseCase(X,
y,
getForestAccuracy,
rfArgs,
"max_depth",
0,
2,
9
)
bgs.showTimeScoreChartAndGraph(max_depth, html=True)Here's what we've got:
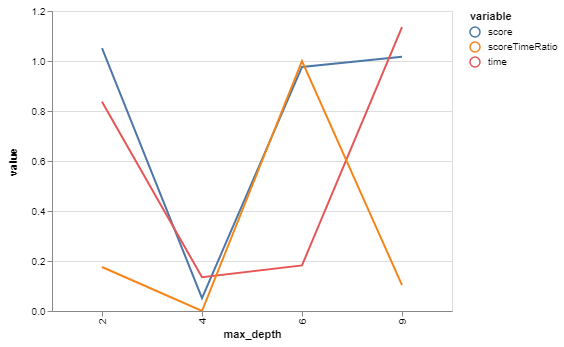
| max_depth | score | time |
|---|---|---|
| 2 | 0.987707 | 0.145360 |
| 9 | 0.987029 | 0.147563 |
| 6 | 0.986247 | 0.140514 |
| 4 | 0.968316 | 0.140164 |
| max_depth | score | time | scoreTimeRatio |
|---|---|---|---|
| 2 | 1.051571 | 0.837377 | 0.175986 |
| 9 | 1.016649 | 1.135158 | 0.103478 |
| 6 | 0.976311 | 0.182516 | 1.000000 |
| 4 | 0.051571 | 0.135158 | 0.000000 |
So, for our purposes, 9 will function as our baseline since that was the biggest depth that it built with default arguments.
Looks like a max_depth of 2 has a slightly higher score than 9, and is slightly faster! Interestingly, it's slightly slower than 4 or 6. Not sure why that is.